Note
Click here to download the full example code
Time-domain CSEM for a resistive cube in a deep marine setting¶
import empymod
import discretize
import pymatsolver
import numpy as np
import SimPEG
from SimPEG import maps
from SimPEG.electromagnetics import time_domain as TDEM
import matplotlib.pyplot as plt
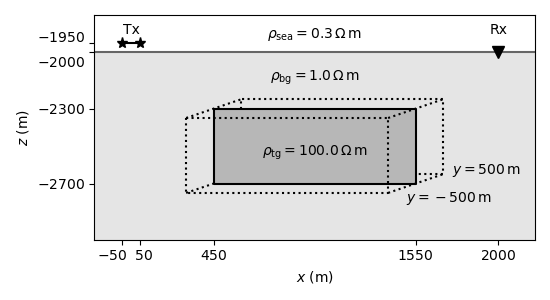
(A) Model¶
fig = plt.figure(figsize=(5.5, 3))
ax = plt.gca()
# Seafloor and background
plt.plot([-200, 2200], [-2000, -2000], "-", c=".4")
bg = plt.Rectangle((-500, -3000), 3000, 1000, facecolor="black", alpha=0.1)
ax.add_patch(bg)
# Plot survey
plt.plot([-50, 50], [-1950, -1950], "*-", ms=8, c="k")
plt.plot(2000, -2000, "v", ms=8, c="k")
plt.text(0, -1900, r"Tx", horizontalalignment="center")
plt.text(2000, -1900, r"Rx", horizontalalignment="center")
# Plot cube
plt.plot([450, 1550, 1550, 450, 450], [-2300, -2300, -2700, -2700, -2300], "k-")
plt.plot([300, 1400, 1400, 300, 300], [-2350, -2350, -2750, -2750, -2350], "k:")
plt.plot([600, 600, 1700, 1700, 1550], [-2300, -2250, -2250, -2650, -2650], "k:")
plt.plot([300, 600], [-2350, -2250], "k:")
plt.plot([1400, 1700], [-2350, -2250], "k:")
plt.plot([300, 450], [-2750, -2700], "k:")
plt.plot([1400, 1700], [-2750, -2650], "k:")
tg = plt.Rectangle((450, -2700), 1100, 400, facecolor="black", alpha=0.2)
ax.add_patch(tg)
# Annotate resistivities
plt.text(
1000, -1925, r"$\rho_\mathrm{sea}=0.3\,\Omega\,$m", horizontalalignment="center"
)
plt.text(
1000, -2150, r"$\rho_\mathrm{bg}=1.0\,\Omega\,$m", horizontalalignment="center"
)
plt.text(
1000, -2550, r"$\rho_\mathrm{tg}=100.0\,\Omega\,$m", horizontalalignment="center"
)
plt.text(1500, -2800, r"$y=-500\,$m", horizontalalignment="left")
plt.text(1750, -2650, r"$y=500\,$m", horizontalalignment="left")
# Ticks and labels
plt.xticks(
[-50, 50, 450, 1550, 2000],
["$-50~$ $~$ $~$", " $~50$", "$450$", "$1550$", "$2000$"],
)
plt.yticks(
[-1950, -2000, -2300, -2700], ["$-1950$\n", "\n$-2000$", "$-2300$", "$-2700$"]
)
plt.xlim([-200, 2200])
plt.ylim([-3000, -1800])
plt.xlabel("$x$ (m)")
plt.ylabel("$z$ (m)")
plt.tight_layout()
plt.show()
Out:
/Users/josephcapriotti/codes/simpeg/examples/06-tdem/plot_fwd_tdem_3d_model.py:71: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
(B) Survey¶
# Source: 100 m x-directed diplole at the origin,
# 50 m above seafloor, src [x1, x2, y1, y2, z1, z2]
src = [-50, 50, 0, 0, -1950, -1950]
# Receiver: x-directed dipole at 2 km on the
# seafloor, rec = [x, y, z, azimuth, dip]
rec = [2000, 0, -2000, 0, 0]
# Times to compute, 0.1 - 10 s, 301 steps
times = np.logspace(-1, 1, 301)
(C) Modelling parameters¶
Check diffusion distances¶
# Get min/max diffusion distances for the two halfspaces.
diff_dist0 = 1261 * np.sqrt(np.r_[times * res_sea, times * res_sea])
diff_dist1 = 1261 * np.sqrt(np.r_[times * res_bg, times * res_bg])
diff_dist2 = 1261 * np.sqrt(np.r_[times * res_tg, times * res_tg])
print("Min/max diffusion distance:")
print(f"- Water :: {diff_dist0.min():8.0f} / {diff_dist0.max():8.0f} m.")
print(f"- Background :: {diff_dist1.min():8.0f} / {diff_dist1.max():8.0f} m.")
print(f"- Target :: {diff_dist2.min():8.0f} / {diff_dist2.max():8.0f} m.")
Out:
Min/max diffusion distance:
- Water :: 218 / 2184 m.
- Background :: 399 / 3988 m.
- Target :: 3988 / 39876 m.
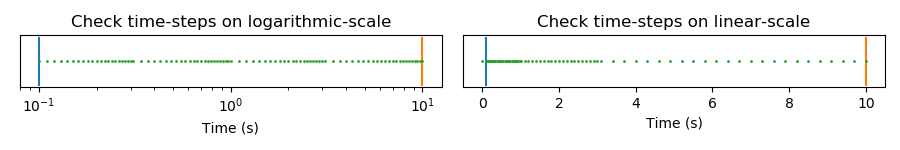
Time-steps¶
# Time steps
time_steps = [1e-1, (1e-2, 21), (3e-2, 23), (1e-1, 21), (3e-1, 23)]
# Create mesh with time steps
ts = discretize.TensorMesh([time_steps]).vectorNx
# Plot them
plt.figure(figsize=(9, 1.5))
# Logarithmic scale
plt.subplot(121)
plt.title("Check time-steps on logarithmic-scale")
plt.plot([times.min(), times.min()], [-1, 1])
plt.plot([times.max(), times.max()], [-1, 1])
plt.plot(ts, ts * 0, ".", ms=2)
plt.yticks([])
plt.xscale("log")
plt.xlabel("Time (s)")
# Linear scale
plt.subplot(122)
plt.title("Check time-steps on linear-scale")
plt.plot([times.min(), times.min()], [-1, 1])
plt.plot([times.max(), times.max()], [-1, 1])
plt.plot(ts, ts * 0, ".", ms=2)
plt.yticks([])
plt.xlabel("Time (s)")
plt.tight_layout()
plt.show()
# Check times with time-steps
print(f"Min/max times : {times.min():.1e} / {times.max():.1e}")
print(f"Min/max timeSteps: {ts[1]:.1e} / {ts[-1]:.1e}")
Out:
/Users/josephcapriotti/codes/simpeg/examples/06-tdem/plot_fwd_tdem_3d_model.py:155: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
Min/max times : 1.0e-01 / 1.0e+01
Min/max timeSteps: 1.0e-01 / 1.0e+01
Create mesh (discretize)¶
# Cell width, number of cells
width = 100
nx = rec[0] // width + 4
ny = 10
nz = 9
# Padding
npadx = 14
npadyz = 12
# Stretching
alpha = 1.3
# Initiate TensorMesh
mesh = discretize.TensorMesh(
[
[(width, npadx, -alpha), (width, nx), (width, npadx, alpha)],
[(width, npadyz, -alpha), (width, ny), (width, npadyz, alpha)],
[(width, npadyz, -alpha), (width, nz), (width, npadyz, alpha)],
],
x0="CCC",
)
# Shift mesh so that
# x=0 is at midpoint of source;
# z=-2000 is at receiver level
mesh.x0[0] += rec[0] // 2 - width / 2
mesh.x0[2] -= nz / 2 * width - seafloor
mesh
| TensorMesh | 58,344 cells | |||||
| MESH EXTENT | CELL WIDTH | FACTOR | ||||
|---|---|---|---|---|---|---|
| dir | nC | min | max | min | max | max |
| x | 52 | -16,878.63 | 18,778.63 | 100.00 | 3,937.38 | 1.30 |
| y | 34 | -10,162.50 | 10,162.50 | 100.00 | 2,329.81 | 1.30 |
| z | 33 | -12,562.50 | 7,662.50 | 100.00 | 2,329.81 | 1.30 |
Check if source and receiver are exactly at x-edges.¶
No requirement; if receiver are exactly on x-edges then no interpolation is required to get the responses (cell centers in x, cell edges in y, z).
print(
f"Rec-{{x;y;z}} :: {rec[0] in np.round(mesh.vectorCCx)!s:>5}; "
f"{rec[1] in np.round(mesh.vectorNy)!s:>5}; "
f"{rec[2] in np.round(mesh.vectorNz)!s:>5}"
)
print(
f"Src-x :: {src[0] in np.round(mesh.vectorCCx)!s:>5}; "
f"{src[1] in np.round(mesh.vectorCCx)!s:>5}"
)
print(
f"Src-y :: {src[2] in np.round(mesh.vectorNy)!s:>5}; "
f"{src[3] in np.round(mesh.vectorNy)!s:>5}"
)
print(
f"Src-z :: {src[4] in np.round(mesh.vectorNz)!s:>5}; "
f"{src[5] in np.round(mesh.vectorNz)!s:>5}"
)
Out:
Rec-{x;y;z} :: True; True; True
Src-x :: False; False
Src-y :: True; True
Src-z :: False; False
Put model on mesh¶
# Background model
mres_bg = np.ones(mesh.nC) * res_sea # Upper halfspace; sea water
mres_bg[mesh.gridCC[:, 2] < seafloor] = res_bg # Lower halfspace; background
# Target model
mres_tg = mres_bg.copy() # Copy background model
target_inds = ( # Find target indices
(mesh.gridCC[:, 0] >= tg_x[0])
& (mesh.gridCC[:, 0] <= tg_x[1])
& (mesh.gridCC[:, 1] >= tg_y[0])
& (mesh.gridCC[:, 1] <= tg_y[1])
& (mesh.gridCC[:, 2] >= tg_z[0])
& (mesh.gridCC[:, 2] <= tg_z[1])
)
mres_tg[target_inds] = res_tg # Target resistivity
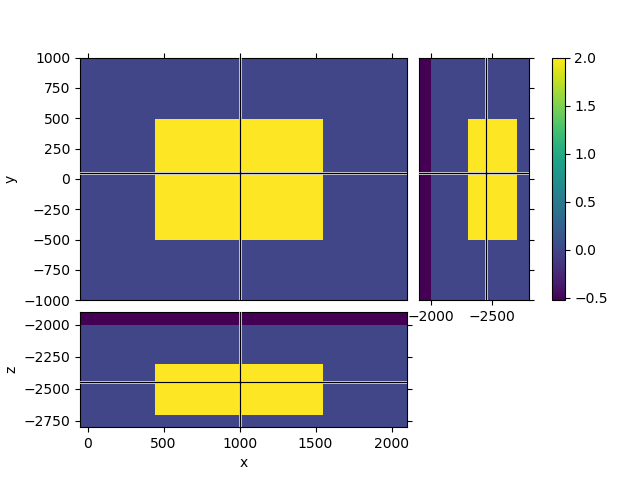
# QC
mesh.plot_3d_slicer(
np.log10(mres_tg),
clim=[np.log10(res_sea), np.log10(res_tg)],
xlim=[-src[0] - 100, rec[0] + 100],
ylim=[-rec[0] / 2, rec[0] / 2],
zlim=[tg_z[0] - 100, seafloor + 100],
)
Out:
/Users/josephcapriotti/opt/anaconda3/envs/py36/lib/python3.6/site-packages/discretize-0.4.11-py3.6-macosx-10.9-x86_64.egg/discretize/View.py:797: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
(D) empymod¶
Compute the 1D background semi-analytically, using 5 points to approximate the 100-m long dipole.
(E) SimPEG¶
Set-up SimPEG-specific parameters.
# Set up the receiver list
rec_list = [
TDEM.Rx.PointElectricField(
orientation="x", times=times, locs=np.array([[*rec[:3]],]),
),
]
# Set up the source list
src_list = [
TDEM.Src.LineCurrent(rxList=rec_list, loc=np.array([[*src[::2]], [*src[1::2]]]),),
]
# Create `Survey`
survey = TDEM.Survey(src_list)
# Define the `Simulation`
prob = TDEM.Simulation3DElectricField(
mesh,
survey=survey,
rhoMap=maps.IdentityMap(mesh),
Solver=pymatsolver.Pardiso,
timeSteps=time_steps,
)
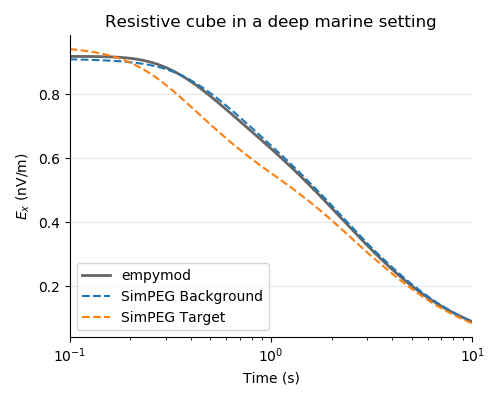
(F) Plots¶
plt.figure(figsize=(5, 4))
ax1 = plt.subplot(111)
plt.title("Resistive cube in a deep marine setting")
plt.plot(times, epm_bg * 1e9, ".4", lw=2, label="empymod")
plt.plot(times, spg_bg * 1e9, "C0--", label="SimPEG Background")
plt.plot(times, spg_tg * 1e9, "C1--", label="SimPEG Target")
plt.ylabel("$E_x$ (nV/m)")
plt.xscale("log")
plt.xlim([0.1, 10])
plt.legend(loc=3)
plt.grid(axis="y", c="0.9")
plt.xlabel("Time (s)")
# Switch off spines
ax1.spines["top"].set_visible(False)
ax1.spines["right"].set_visible(False)
plt.tight_layout()
plt.show()
Out:
/Users/josephcapriotti/codes/simpeg/examples/06-tdem/plot_fwd_tdem_3d_model.py:345: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
empymod.Report([SimPEG, discretize, pymatsolver])
| Tue May 26 19:12:42 2020 MDT | |||||
| OS | Darwin | CPU(s) | 8 | Machine | x86_64 |
| Architecture | 64bit | Environment | Python | ||
| Python 3.6.10 |Anaconda, Inc.| (default, May 7 2020, 23:06:31) [GCC 4.2.1 Compatible Clang 4.0.1 (tags/RELEASE_401/final)] | |||||
| SimPEG | 0.14.0b2 | discretize | 0.4.11 | pymatsolver | 0.1.2 |
| numpy | 1.18.1 | scipy | 1.4.1 | numba | 0.49.1 |
| empymod | 2.0.0 | IPython | 7.13.0 | matplotlib | 3.1.3 |
| Intel(R) Math Kernel Library Version 2019.0.4 Product Build 20190411 for Intel(R) 64 architecture applications | |||||
Total running time of the script: ( 3 minutes 27.584 seconds)