Note
Click here to download the full example code
Forward Simulation of Gravity Anomaly Data on a Tensor Mesh¶
Here we use the module SimPEG.potential_fields.gravity to predict gravity anomaly data for a synthetic density contrast model. The simulation is carried out on a tensor mesh. For this tutorial, we focus on the following:
How to create gravity surveys
How to predict gravity anomaly data for a density contrast model
How to include surface topography
The units of the density contrast model and resulting data
Import Modules¶
import numpy as np
from scipy.interpolate import LinearNDInterpolator
import matplotlib as mpl
import matplotlib.pyplot as plt
import os
from discretize import TensorMesh
from discretize.utils import mkvc
from SimPEG.utils import plot2Ddata, model_builder, surface2ind_topo
from SimPEG import maps
from SimPEG.potential_fields import gravity
save_file = False
# sphinx_gallery_thumbnail_number = 2
Defining Topography¶
Surface topography is defined as an (N, 3) numpy array. We create it here but the topography could also be loaded from a file.
Defining the Survey¶
Here, we define survey that will be used for the forward simulation. Gravity surveys are simple to create. The user only needs an (N, 3) array to define the xyz locations of the observation locations, and a list of field components which are to be measured.
# Define the observation locations as an (N, 3) numpy array or load them.
x = np.linspace(-80.0, 80.0, 17)
y = np.linspace(-80.0, 80.0, 17)
x, y = np.meshgrid(x, y)
x, y = mkvc(x.T), mkvc(y.T)
fun_interp = LinearNDInterpolator(np.c_[x_topo, y_topo], z_topo)
z = fun_interp(np.c_[x, y]) + 5.0
receiver_locations = np.c_[x, y, z]
# Define the component(s) of the field we want to simulate as strings within
# a list. Here we simulate only the vertical component of gravity anomaly.
components = ["gz"]
# Use the observation locations and components to define the receivers. To
# simulate data, the receivers must be defined as a list.
receiver_list = gravity.receivers.Point(receiver_locations, components=components)
receiver_list = [receiver_list]
# Defining the source field.
source_field = gravity.sources.SourceField(receiver_list=receiver_list)
# Defining the survey
survey = gravity.survey.Survey(source_field)
Defining a Tensor Mesh¶
Here, we create the tensor mesh that will be used to predict gravity anomaly data.
Density Contrast Model and Mapping on Tensor Mesh¶
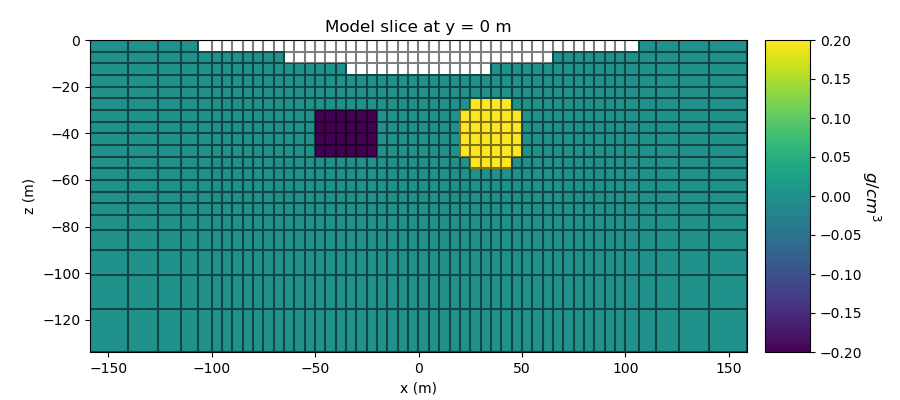
Here, we create the density contrast model that will be used to predict gravity anomaly data and the mapping from the model to the mesh. The model consists of a less dense block and a more dense sphere.
# Define density contrast values for each unit in g/cc
background_density = 0.0
block_density = -0.2
sphere_density = 0.2
# Find the indices for the active mesh cells (e.g. cells below surface)
ind_active = surface2ind_topo(mesh, xyz_topo)
# Define mapping from model to active cells. The model consists of a value for
# each cell below the Earth's surface.
nC = int(ind_active.sum())
model_map = maps.IdentityMap(nP=nC)
# Define model. Models in SimPEG are vector arrays.
model = background_density * np.ones(nC)
# You could find the indicies of specific cells within the model and change their
# value to add structures.
ind_block = (
(mesh.gridCC[ind_active, 0] > -50.0)
& (mesh.gridCC[ind_active, 0] < -20.0)
& (mesh.gridCC[ind_active, 1] > -15.0)
& (mesh.gridCC[ind_active, 1] < 15.0)
& (mesh.gridCC[ind_active, 2] > -50.0)
& (mesh.gridCC[ind_active, 2] < -30.0)
)
model[ind_block] = block_density
# You can also use SimPEG utilities to add structures to the model more concisely
ind_sphere = model_builder.getIndicesSphere(np.r_[35.0, 0.0, -40.0], 15.0, mesh.gridCC)
ind_sphere = ind_sphere[ind_active]
model[ind_sphere] = sphere_density
# Plot Density Contrast Model
fig = plt.figure(figsize=(9, 4))
plotting_map = maps.InjectActiveCells(mesh, ind_active, np.nan)
ax1 = fig.add_axes([0.1, 0.12, 0.73, 0.78])
mesh.plotSlice(
plotting_map * model,
normal="Y",
ax=ax1,
ind=int(mesh.nCy / 2),
grid=True,
clim=(np.min(model), np.max(model)),
pcolorOpts={"cmap": "viridis"},
)
ax1.set_title("Model slice at y = 0 m")
ax1.set_xlabel("x (m)")
ax1.set_ylabel("z (m)")
ax2 = fig.add_axes([0.85, 0.12, 0.05, 0.78])
norm = mpl.colors.Normalize(vmin=np.min(model), vmax=np.max(model))
cbar = mpl.colorbar.ColorbarBase(
ax2, norm=norm, orientation="vertical", cmap=mpl.cm.viridis
)
cbar.set_label("$g/cm^3$", rotation=270, labelpad=15, size=12)
plt.show()
Simulation: Gravity Anomaly Data on Tensor Mesh¶
Here we demonstrate how to predict gravity anomaly data using the integral formulation.
# Define the forward simulation. By setting the 'store_sensitivities' keyword
# argument to "forward_only", we simulate the data without storing the sensitivities
simulation = gravity.simulation.Simulation3DIntegral(
survey=survey,
mesh=mesh,
rhoMap=model_map,
actInd=ind_active,
store_sensitivities="forward_only",
)
# Compute predicted data for some model
dpred = simulation.dpred(model)
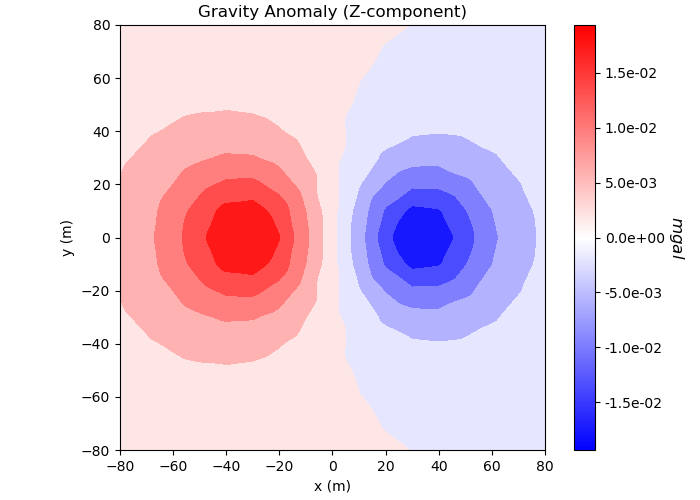
# Plot
fig = plt.figure(figsize=(7, 5))
ax1 = fig.add_axes([0.1, 0.1, 0.75, 0.85])
plot2Ddata(receiver_list[0].locations, dpred, ax=ax1, contourOpts={"cmap": "bwr"})
ax1.set_title("Gravity Anomaly (Z-component)")
ax1.set_xlabel("x (m)")
ax1.set_ylabel("y (m)")
ax2 = fig.add_axes([0.82, 0.1, 0.03, 0.85])
norm = mpl.colors.Normalize(vmin=-np.max(np.abs(dpred)), vmax=np.max(np.abs(dpred)))
cbar = mpl.colorbar.ColorbarBase(
ax2, norm=norm, orientation="vertical", cmap=mpl.cm.bwr, format="%.1e"
)
cbar.set_label("$mgal$", rotation=270, labelpad=15, size=12)
plt.show()
Optional: Exporting Results¶
Write the data, topography and true model
if save_file:
dir_path = os.path.dirname(gravity.__file__).split(os.path.sep)[:-3]
dir_path.extend(["tutorials", "assets", "gravity"])
dir_path = os.path.sep.join(dir_path) + os.path.sep
fname = dir_path + "gravity_topo.txt"
np.savetxt(fname, np.c_[xyz_topo], fmt="%.4e")
maximum_anomaly = np.max(np.abs(dpred))
noise = 0.01 * maximum_anomaly * np.random.rand(len(dpred))
fname = dir_path + "gravity_data.obs"
np.savetxt(fname, np.c_[receiver_locations, dpred + noise], fmt="%.4e")
output_model = plotting_map * model
output_model[np.isnan(output_model)] = 0.0
fname = dir_path + "true_model.txt"
np.savetxt(fname, output_model, fmt="%.4e")
Total running time of the script: ( 0 minutes 14.312 seconds)