Note
Click here to download the full example code
DC Resistivity Forward Simulation in 2.5D¶
Here we use the module SimPEG.electromagnetics.static.resistivity to predict DC resistivity data. In this tutorial, we focus on the following:
How to define the survey
How to define the forward simulation
How to predict DC resistivity data for a synthetic resistivity model
How to include surface topography
The units of the model and resulting data
Import modules¶
from discretize import TreeMesh
from discretize.utils import mkvc, refine_tree_xyz
from SimPEG.utils import model_builder, surface2ind_topo
from SimPEG import maps, data
from SimPEG.electromagnetics.static import resistivity as dc
from SimPEG.electromagnetics.static.utils.static_utils import (
generate_dcip_survey_line,
plot_pseudoSection,
)
import os
import numpy as np
import matplotlib as mpl
import matplotlib.pyplot as plt
try:
from pymatsolver import Pardiso as Solver
except ImportError:
from SimPEG import SolverLU as Solver
save_file = False
# sphinx_gallery_thumbnail_number = 2
Defining Topography¶
Here we define surface topography as an (N, 3) numpy array. Topography could also be loaded from a file. In our case, our survey takes place in a valley that runs North-South.
Create Dipole-Dipole Survey¶
Here we define a single EW survey line that uses a dipole-dipole configuration. For the source, we must define the AB electrode locations. For the receivers we must define the MN electrode locations. Instead of creating the survey from scratch (see 1D example), we will use the generat_dcip_survey_line utility.
# Define survey line parameters
survey_type = "dipole-dipole"
data_type = "volt"
end_locations = np.r_[-400.0, 400]
station_separation = 50.0
dipole_separation = 25.0
n = 8
# Generate DC survey line
survey = generate_dcip_survey_line(
survey_type,
data_type,
end_locations,
xyz_topo,
station_separation,
dipole_separation,
n,
dim_flag="2.5D",
sources_only=False,
)
Create OcTree Mesh¶
Here, we create the OcTree mesh that will be used to predict both DC resistivity and IP data.
dh = 10.0 # base cell width
dom_width_x = 2400.0 # domain width x
dom_width_z = 1200.0 # domain width z
nbcx = 2 ** int(np.round(np.log(dom_width_x / dh) / np.log(2.0))) # num. base cells x
nbcz = 2 ** int(np.round(np.log(dom_width_z / dh) / np.log(2.0))) # num. base cells z
# Define the base mesh
hx = [(dh, nbcx)]
hz = [(dh, nbcz)]
mesh = TreeMesh([hx, hz], x0="CN")
# Mesh refinement based on topography
mesh = refine_tree_xyz(
mesh, xyz_topo[:, [0, 2]], octree_levels=[1], method="surface", finalize=False
)
# Mesh refinement near transmitters and receivers. First we need to obtain the
# set of unique electrode locations.
electrode_locations = np.c_[
survey.locations_a, survey.locations_b, survey.locations_m, survey.locations_n
]
unique_locations = np.unique(
np.reshape(electrode_locations, (4 * survey.nD, 2)), axis=0
)
mesh = refine_tree_xyz(
mesh, unique_locations, octree_levels=[2, 4], method="radial", finalize=False
)
# Refine core mesh region
xp, zp = np.meshgrid([-800.0, 800.0], [-800.0, 0.0])
xyz = np.c_[mkvc(xp), mkvc(zp)]
mesh = refine_tree_xyz(mesh, xyz, octree_levels=[0, 2, 2], method="box", finalize=False)
mesh.finalize()
Create Conductivity Model and Mapping for OcTree Mesh¶
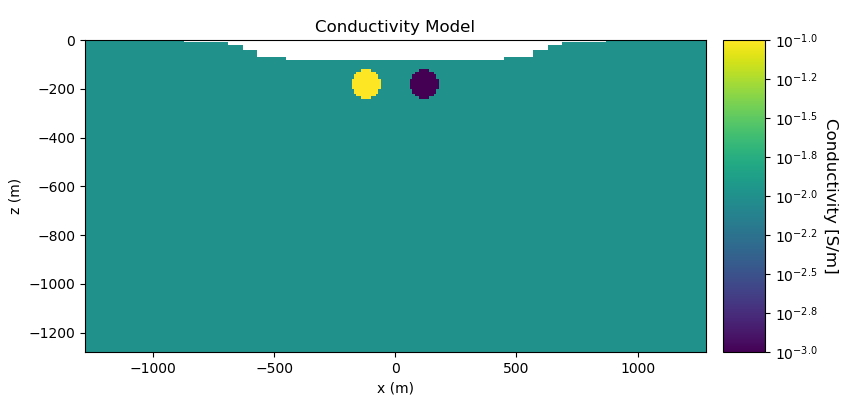
Here we define the conductivity model that will be used to predict DC resistivity data. The model consists of a conductive sphere and a resistive sphere within a moderately conductive background. Note that you can carry through this work flow with a resistivity model if desired.
# Define conductivity model in S/m (or resistivity model in Ohm m)
air_conductivity = 1e-8
background_conductivity = 1e-2
conductor_conductivity = 1e-1
resistor_conductivity = 1e-3
# Find active cells in forward modeling (cell below surface)
ind_active = surface2ind_topo(mesh, xyz_topo[:, [0, 2]])
# Define mapping from model to active cells
nC = int(ind_active.sum())
conductivity_map = maps.InjectActiveCells(mesh, ind_active, air_conductivity)
# Define model
conductivity_model = background_conductivity * np.ones(nC)
ind_conductor = model_builder.getIndicesSphere(np.r_[-120.0, -180.0], 60.0, mesh.gridCC)
ind_conductor = ind_conductor[ind_active]
conductivity_model[ind_conductor] = conductor_conductivity
ind_resistor = model_builder.getIndicesSphere(np.r_[120.0, -180.0], 60.0, mesh.gridCC)
ind_resistor = ind_resistor[ind_active]
conductivity_model[ind_resistor] = resistor_conductivity
# Plot Conductivity Model
fig = plt.figure(figsize=(8.5, 4))
plotting_map = maps.InjectActiveCells(mesh, ind_active, np.nan)
log_mod = np.log10(conductivity_model)
ax1 = fig.add_axes([0.1, 0.12, 0.73, 0.78])
mesh.plotImage(
plotting_map * log_mod,
ax=ax1,
grid=False,
clim=(np.log10(resistor_conductivity), np.log10(conductor_conductivity)),
pcolorOpts={"cmap": "viridis"},
)
ax1.set_title("Conductivity Model")
ax1.set_xlabel("x (m)")
ax1.set_ylabel("z (m)")
ax2 = fig.add_axes([0.85, 0.12, 0.05, 0.78])
norm = mpl.colors.Normalize(
vmin=np.log10(resistor_conductivity), vmax=np.log10(conductor_conductivity)
)
cbar = mpl.colorbar.ColorbarBase(
ax2, norm=norm, cmap=mpl.cm.viridis, orientation="vertical", format="$10^{%.1f}$"
)
cbar.set_label("Conductivity [S/m]", rotation=270, labelpad=15, size=12)
Project Survey to Discretized Topography¶
It is important that electrodes are not model as being in the air. Even if the electrodes are properly located along surface topography, they may lie above the discretized topography. This step is carried out to ensure all electrodes like on the discretized surface.
survey.drape_electrodes_on_topography(mesh, ind_active, option="top")
Predict DC Resistivity Data¶
Here we predict DC resistivity data. If the keyword argument sigmaMap is defined, the simulation will expect a conductivity model. If the keyword argument rhoMap is defined, the simulation will expect a resistivity model.
simulation = dc.simulation_2d.Simulation2DNodal(
mesh, survey=survey, sigmaMap=conductivity_map, Solver=Solver
)
# Predict the data by running the simulation. The data are the raw voltage in
# units of volts.
dpred = simulation.dpred(conductivity_model)
# Define a data object (required for pseudo-section plot)
data_obj = data.Data(survey, dobs=dpred)
# Plot apparent conductivity pseudo-section
fig = plt.figure(figsize=(12, 5))
ax1 = fig.add_axes([0.05, 0.05, 0.8, 0.9])
plot_pseudoSection(
data_obj,
ax=ax1,
survey_type="dipole-dipole",
data_type="appConductivity",
space_type="half-space",
scale="log",
pcolorOpts={"cmap": "viridis"},
)
ax1.set_title("Apparent Conductivity [S/m]")
plt.show()
![Apparent Conductivity [S/m]](../../../_images/sphx_glr_plot_fwd_2_dcr2d_002.png)
Out:
/Users/josephcapriotti/codes/simpeg/tutorials/05-dcr/plot_fwd_2_dcr2d.py:253: UserWarning: Matplotlib is currently using agg, which is a non-GUI backend, so cannot show the figure.
plt.show()
Optional: Write out dpred¶
Write DC resistivity data, topography and true model
if save_file:
dir_path = os.path.dirname(dc.__file__).split(os.path.sep)[:-4]
dir_path.extend(["tutorials", "assets", "dcr2d"])
dir_path = os.path.sep.join(dir_path) + os.path.sep
# Add 5% Gaussian noise to each datum
noise = 0.05 * dpred * np.random.rand(len(dpred))
# Write out data at their original electrode locations (not shifted)
data_array = np.c_[electrode_locations, dpred + noise]
fname = dir_path + "dc_data.obs"
np.savetxt(fname, data_array, fmt="%.4e")
fname = dir_path + "true_conductivity.txt"
np.savetxt(fname, conductivity_map * conductivity_model, fmt="%.4e")
fname = dir_path + "xyz_topo.txt"
np.savetxt(fname, xyz_topo, fmt="%.4e")
Total running time of the script: ( 0 minutes 3.494 seconds)